Implementing linear regression in Python
Contents
Implementing linear regression in Python¶
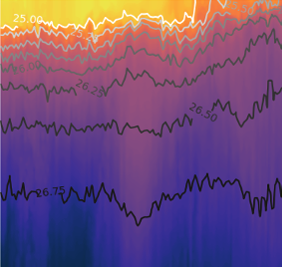
Example: 2007 West Coast Ocean Acidification Cruise¶
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from scipy import stats
import statsmodels.formula.api as smf
import pingouin as pg
import PyCO2SYS as pyco2
/Users/tomconnolly/programs/miniconda3/envs/data-book/lib/python3.8/site-packages/outdated/utils.py:14: OutdatedPackageWarning: The package pingouin is out of date. Your version is 0.5.0, the latest is 0.5.1.
Set the environment variable OUTDATED_IGNORE=1 to disable these warnings.
return warn(
Load data¶
Load 2007 data¶
filename07 = 'wcoa_cruise_2007/32WC20070511.exc.csv'
df07 = pd.read_csv(filename07,header=29,na_values=-999,parse_dates=[[6,7]])
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/var/folders/67/t9t76vyd78qgn_kwjr1_76yr0000gn/T/ipykernel_74292/425823264.py in <module>
1 filename07 = 'wcoa_cruise_2007/32WC20070511.exc.csv'
----> 2 df07 = pd.read_csv(filename07,header=29,na_values=-999,parse_dates=[[6,7]])
~/programs/miniconda3/envs/data-book/lib/python3.8/site-packages/pandas/util/_decorators.py in wrapper(*args, **kwargs)
309 stacklevel=stacklevel,
310 )
--> 311 return func(*args, **kwargs)
312
313 return wrapper
~/programs/miniconda3/envs/data-book/lib/python3.8/site-packages/pandas/io/parsers/readers.py in read_csv(filepath_or_buffer, sep, delimiter, header, names, index_col, usecols, squeeze, prefix, mangle_dupe_cols, dtype, engine, converters, true_values, false_values, skipinitialspace, skiprows, skipfooter, nrows, na_values, keep_default_na, na_filter, verbose, skip_blank_lines, parse_dates, infer_datetime_format, keep_date_col, date_parser, dayfirst, cache_dates, iterator, chunksize, compression, thousands, decimal, lineterminator, quotechar, quoting, doublequote, escapechar, comment, encoding, encoding_errors, dialect, error_bad_lines, warn_bad_lines, on_bad_lines, delim_whitespace, low_memory, memory_map, float_precision, storage_options)
584 kwds.update(kwds_defaults)
585
--> 586 return _read(filepath_or_buffer, kwds)
587
588
~/programs/miniconda3/envs/data-book/lib/python3.8/site-packages/pandas/io/parsers/readers.py in _read(filepath_or_buffer, kwds)
480
481 # Create the parser.
--> 482 parser = TextFileReader(filepath_or_buffer, **kwds)
483
484 if chunksize or iterator:
~/programs/miniconda3/envs/data-book/lib/python3.8/site-packages/pandas/io/parsers/readers.py in __init__(self, f, engine, **kwds)
809 self.options["has_index_names"] = kwds["has_index_names"]
810
--> 811 self._engine = self._make_engine(self.engine)
812
813 def close(self):
~/programs/miniconda3/envs/data-book/lib/python3.8/site-packages/pandas/io/parsers/readers.py in _make_engine(self, engine)
1038 )
1039 # error: Too many arguments for "ParserBase"
-> 1040 return mapping[engine](self.f, **self.options) # type: ignore[call-arg]
1041
1042 def _failover_to_python(self):
~/programs/miniconda3/envs/data-book/lib/python3.8/site-packages/pandas/io/parsers/c_parser_wrapper.py in __init__(self, src, **kwds)
49
50 # open handles
---> 51 self._open_handles(src, kwds)
52 assert self.handles is not None
53
~/programs/miniconda3/envs/data-book/lib/python3.8/site-packages/pandas/io/parsers/base_parser.py in _open_handles(self, src, kwds)
220 Let the readers open IOHandles after they are done with their potential raises.
221 """
--> 222 self.handles = get_handle(
223 src,
224 "r",
~/programs/miniconda3/envs/data-book/lib/python3.8/site-packages/pandas/io/common.py in get_handle(path_or_buf, mode, encoding, compression, memory_map, is_text, errors, storage_options)
700 if ioargs.encoding and "b" not in ioargs.mode:
701 # Encoding
--> 702 handle = open(
703 handle,
704 ioargs.mode,
FileNotFoundError: [Errno 2] No such file or directory: 'wcoa_cruise_2007/32WC20070511.exc.csv'
df07.keys()
Index(['DATE_TIME', 'EXPOCODE', 'SECT_ID', 'SAMPNO', 'LINE', 'STNNBR',
'CASTNO', 'LATITUDE', 'LONGITUDE', 'BOT_DEPTH', 'BTLNBR',
'BTLNBR_FLAG_W', 'CTDPRS', 'CTDTMP', 'CTDSAL', 'CTDSAL_FLAG_W',
'SALNTY', 'SALNTY_FLAG_W', 'CTDOXY', 'CTDOXY_FLAG_W', 'OXYGEN',
'OXYGEN_FLAG_W', 'SILCAT', 'SILCAT_FLAG_W', 'NITRAT', 'NITRAT_FLAG_W',
'NITRIT', 'NITRIT_FLAG_W', 'PHSPHT', 'PHSPHT_FLAG_W', 'TCARBN',
'TCARBN_FLAG_W', 'ALKALI', 'ALKALI_FLAG_W'],
dtype='object')
Linear regression: four methods in Python¶
Create \(x\) and \(y\) variables.
x = df07['PHSPHT']
y = df07['NITRAT']
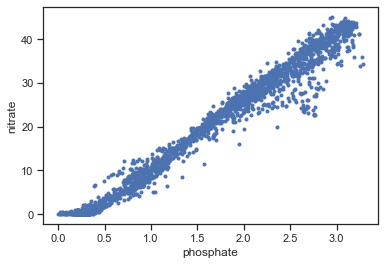
Plot data.
plt.figure()
plt.plot(x,y,'.')
plt.xlabel('phosphate')
plt.ylabel('nitrate')
Text(0, 0.5, 'nitrate')
Create a subset where both variables have finite values.
ii = np.isfinite(x+y)
ii
0 True
1 True
2 True
3 True
4 True
...
2343 True
2344 True
2345 True
2346 True
2347 True
Length: 2348, dtype: bool
Method 1: Scipy¶
result = stats.linregress(x[ii],y[ii])
result
LinregressResult(slope=14.740034517902107, intercept=-3.9325720551997954, rvalue=0.9860645445968036, pvalue=0.0, stderr=0.05292378356970245, intercept_stderr=0.10258209230911518)
result.slope
14.740034517902107
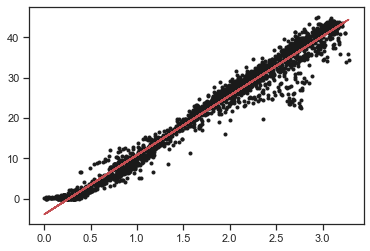
Exercise: Draw the regression line with the data
plt.figure()
plt.plot(x,y,'k.')
plt.plot(x,result.slope*x+result.intercept,'r-')
[<matplotlib.lines.Line2D at 0x7f8be85239d0>]
Method 2: statsmodels¶
Ordinary least squares fit using statsmodels.
smres = smf.ols('NITRAT ~ PHSPHT',df07).fit()
smres.summary()
| Dep. Variable: | NITRAT | R-squared: | 0.972 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.972 |
| Method: | Least Squares | F-statistic: | 7.757e+04 |
| Date: | Tue, 22 Feb 2022 | Prob (F-statistic): | 0.00 |
| Time: | 21:22:25 | Log-Likelihood: | -4993.9 |
| No. Observations: | 2210 | AIC: | 9992. |
| Df Residuals: | 2208 | BIC: | 1.000e+04 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -3.9326 | 0.103 | -38.336 | 0.000 | -4.134 | -3.731 |
| PHSPHT | 14.7400 | 0.053 | 278.514 | 0.000 | 14.636 | 14.844 |
| Omnibus: | 874.728 | Durbin-Watson: | 0.269 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 5147.310 |
| Skew: | -1.766 | Prob(JB): | 0.00 |
| Kurtosis: | 9.589 | Cond. No. | 4.90 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Method 3: pingouin¶
pg.linear_regression(x[ii],y[ii])
| names | coef | se | T | pval | r2 | adj_r2 | CI[2.5%] | CI[97.5%] | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Intercept | -3.932572 | 0.102582 | -38.335853 | 6.540950e-247 | 0.972323 | 0.972311 | -4.133740 | -3.731405 |
| 1 | PHSPHT | 14.740035 | 0.052924 | 278.514375 | 0.000000e+00 | 0.972323 | 0.972311 | 14.636249 | 14.843820 |